Comparative study to find mutational pattern in thermophilic bacteria
Creating thermostable enzymes, remains one of the essential requirements in protein engineering, but there is a difficulty in designing a general method, because benefit from mutations observed in evolution studies is difficult to rationalize in molecular perspective as mutations occur far from active site of enzymes. My objective with this study was to gain more insights in the molecular evolution of Thermophiles.
Docking studies of molecules with PfA-M1 alanyl-aminopeptidase
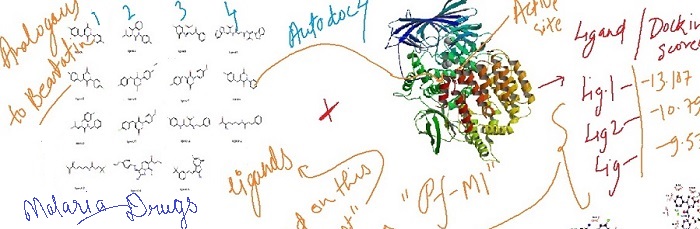
M1 alanyl-aminopeptidase (PfA-M1) is a potential target for the treatment of malaria. With increase in resistance against most antimalarial drug it is now imperative to find new drug target M1 alanyl aminopeptidase. . The PfA- M1 crystal structure was retrieved from protein data bank. Fifteen (15) ligands, obtained from screening were docked with PfA-M1 using AUTODOCK. The binding affinity of these ligands was compared with that of Bestatin. Interestingly, some of the compounds showed very similar or better docking scores as compared to bestatin which is a natural inhibitor of m1 fa. These selected compounds can be further studied for developing potential inhibitors against PfA-M1.



